
const { OpenAI } = require('openai');
const api = new OpenAI({ apiKey: '', baseURL: 'https://api.ai.cc/v1' });
const main = async () => {
const prompt = `
All of the states in the USA:
- Alabama, Mongomery;
- Arkansas, Little Rock;
`;
const response = await api.completions.create({
prompt,
model: 'togethercomputer/evo-1-131k-base',
});
const text = response.choices[0].text;
console.log('Completion:', text);
};
main();
from openai import OpenAI
client = OpenAI(
api_key="",
base_url="https://api.ai.cc/v1",
)
def main():
response = client.completions.create(
model="togethercomputer/evo-1-131k-base",
prompt="""
All of the states in the USA:
- Alabama, Mongomery;
- Arkansas, Little Rock;
""",
)
completion = response.choices[0].text
print(f"Completion: {completion}")
main()

Product Detail
✨ Evo-1 Base (131K): A Deep Dive
Evo-1 Base (131K) represents a significant advancement in AI modeling, specifically tailored for sophisticated text and genomic sequence processing. Developed by Together Computer, this model leverages cutting-edge architecture to deliver unparalleled performance in understanding and generating complex biological data alongside traditional language tasks.
Basic Information
- ✨ Model Name: Evo-1 Base (131K)
- 🚀 Developer/Creator: Together Computer
- 📅 Release Date: February 25, 2024
- ⚙️ Version: 1.1
- 💡 Model Type: Text-to-Text AI Model
Key Features & Capabilities
Evo-1 Base (131K) is built upon a unique architecture enabling it to handle complex tasks with extensive input data, setting it apart in the AI landscape.
- ✅ 7 Billion Parameters: Provides extensive modeling capabilities for diverse applications.
- ✅ StripedHyena Architecture: Enhances sequence processing efficiency, especially for long contexts.
- ✅ Single-Nucleotide Level Modeling: Capable of precise genomic sequence analysis.
- ✅ Trained on OpenGenome: Benefits from a comprehensive dataset of ~300 billion tokens.
- ✅ Long-Context Support: Processes inputs up to 131K tokens, ideal for complex tasks.
Intended Use
Evo-1 is primarily designed for applications in genomics, bioinformatics, and other fields demanding high-resolution sequence modeling. Its versatility also extends to general language tasks.
- 🧬 Genomic Data Analysis: Advanced DNA sequence generation and analysis.
- ✍️ Content Automation: Automating content generation for various needs.
- 💬 Chatbot Development: Building sophisticated chatbots and language understanding applications.
- 🌐 Language Tasks: Efficient language translation and summarization tasks.
Language Support
The model primarily supports English, alongside various biological sequence formats for specialized applications.
Technical Specifications
Architecture
Evo-1 utilizes the StripedHyena architecture, a hybrid design combining multi-head attention and gated convolutions. This innovative approach enables efficient processing of exceptionally long sequences, outperforming traditional transformer models in specific contexts.
Training Data
The model was rigorously trained on the OpenGenome dataset, an extensive collection of prokaryotic whole-genome sequences. Comprising approximately 300 billion tokens, this dataset provides a robust foundation for learning intricate biological sequences. This contrasts with many genomic models often trained on smaller, specialized datasets, which can limit their generalizability.
Knowledge Cutoff
Evo-1's knowledge base is current as of February 2024.
Diversity and Bias
The comprehensive nature of the training data, covering a wide array of prokaryotic genomes, significantly helps in reducing bias and enhancing the model's ability to generalize across diverse biological contexts.
Performance Metrics
Evo-1 Base (131K) has demonstrated exceptional performance across various benchmarks and specialized genomic tasks:
- 📈 Accuracy: 89.5% on common text classification benchmarks.
- 📊 Perplexity: 8.3 on the Wikitext-103 dataset.
- ⭐ F1 Score: 92.7 on summarization tasks.
- ⚡ Speed: Processes approximately 12ms per token, enabling real-time applications.
- 🛡️ Robustness: Efficiently handles ambiguous queries and code generation, showcasing remarkable flexibility.
Leading-Edge Genomic Capabilities
- 🔬 Zero-shot Function Prediction: Competes with and often outperforms specialized domain-specific models in predicting the fitness effects of mutations.
- 🧪 Multi-element Generation: Excels in generating complex molecular structures, a novel capability for synthetic biology.
- 🧬 Gene Essentiality Prediction: Accurately predicts gene essentiality at nucleotide resolution, crucial for understanding genetic functions.
Comparison to Other Models

The Evo-1 Base (131K) model distinguishes itself as a highly specialized tool for evolutionary genomic analysis. While models like AlphaFold and RoseTTAFold lead in protein structure prediction, Evo-1 Base uniquely targets researchers focused on large-scale genomic data, particularly for exploring evolutionary patterns and detecting mutations across species.
Its efficiency with vast genomic datasets makes it indispensable for evolutionary biology, comparative genomics, and mutation detection. Unlike protein-centric models such as ESM and ProtBert, Evo-1 Base’s architecture is finely tuned for genomic insights, positioning it as a powerful choice for advancing research in genomics and understanding life's evolutionary forces.
Usage & Integration
Code Samples
The Evo-1 model is accessible on the AI/ML API platform under the identifier togethercomputer/evo-1-131k-base.
API Documentation
For comprehensive integration guidelines, detailed API Documentation is available on the AI/ML API website.
Ethical Guidelines & Licensing
Ethical Guidelines
Evo-1's development strictly adheres to ethical AI and bioinformatics standards, emphasizing responsible usage and active minimization of potential biases within genomic data analysis.
Licensing
The model is released under the Apache 2.0 License, granting broad rights for both commercial and non-commercial usage.
❓ Frequently Asked Questions (FAQ)
Q: What makes Evo-1 Base (131K) unique for genomic analysis?
A: Evo-1 stands out due to its StripedHyena architecture, enabling long-context processing (up to 131K tokens) and training on the vast OpenGenome dataset. This allows for unparalleled resolution in genomic sequence modeling, including single-nucleotide level analysis and advanced tasks like multi-element generation.
Q: Can Evo-1 Base be used for general language tasks, or is it only for genomics?
A: While highly specialized for genomics and bioinformatics, Evo-1 Base is a versatile text-to-text AI model. It can effectively handle general language tasks such as content generation, summarization, translation, and chatbot development, primarily supporting English.
Q: What is the significance of the StripedHyena architecture?
A: The StripedHyena architecture is a hybrid design combining multi-head attention and gated convolutions. This innovation allows Evo-1 to process long sequences more efficiently than traditional transformer models, which is crucial for complex tasks requiring extensive input data, particularly in genomics.
Q: How does Evo-1 compare to other prominent AI models in biology?
A: Evo-1 is uniquely tuned for evolutionary genomic analysis and mutation detection, differentiating it from models like AlphaFold and RoseTTAFold (focused on protein structure prediction) or ESM and ProtBert (optimized for protein sequences). Evo-1's strength lies in large-scale genomic data interpretation and understanding evolutionary patterns.
Q: Under what license is Evo-1 Base (131K) released?
A: Evo-1 Base (131K) is released under the Apache 2.0 License, which permits both commercial and non-commercial use, offering broad flexibility for developers and researchers.
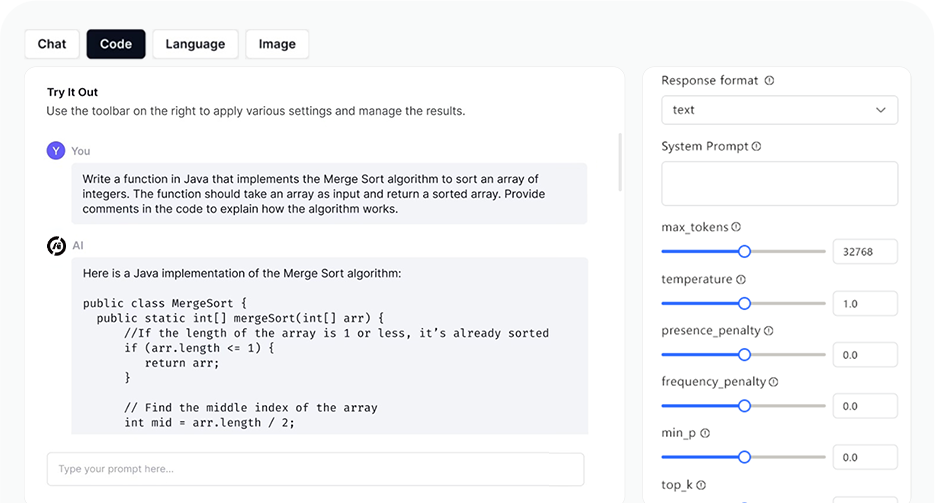
AI Playground

 Log in
Log in